
Powered by software container technology (Docker) and a workflow manager (Nextflow), the DEBBIE pipeline automatically and continuously retrieves research abstracts, filters them according to relevance using the DEBBIE_BioBERT model, annotates concepts using the Biomaterials Annotator, and stores the information within a document-oriented database named the Database of Experimental Biomaterials and their Biological Effect (DEBBIE).
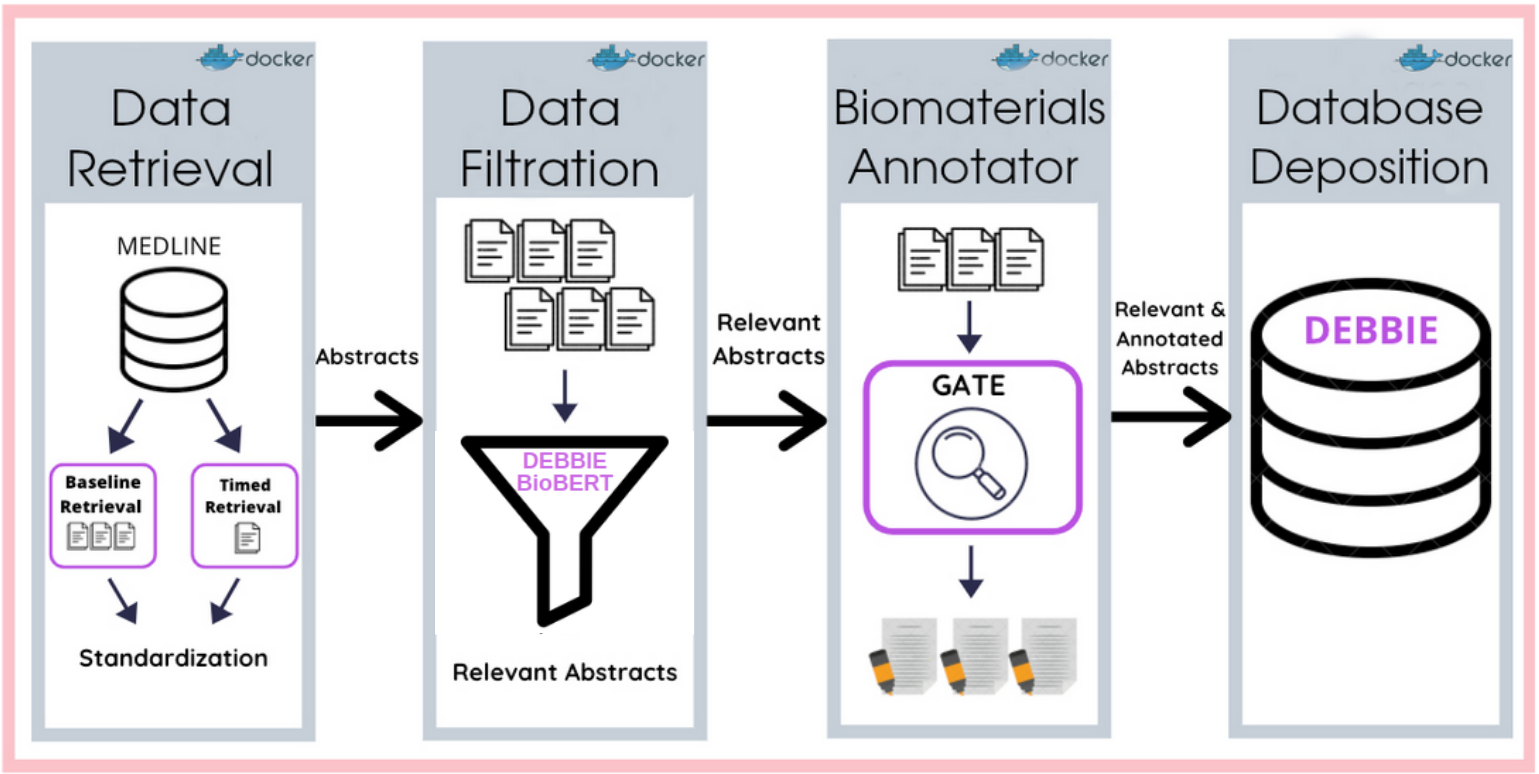
As a starting point for the gathering of relevant biomaterials abstracts, the following PubMed search query is executed: ((((((((Biomedical and dental materials[MeSH Terms]) OR (Prostheses and implants[MeSH Terms])) OR (Materials testing[MeSH Terms])) OR (Tissue engineering[MeSH Terms])) OR (Tissue scaffolds[MeSH Terms])) OR (Equipment safety[MeSH Terms])) OR (Medical device recalls[MeSH Terms])) OR (Biomaterials)) OR (Cell scaffolds).
The DEBBIE_BioBERT model performs multiclass classification of abstracts to determine if they are relevant (either clinical or non-clinical studies) or not relevant to the field of biomaterials. The DEBBIE_BioBERT model was developed using Transformers, which is the state-of-the-art in NLP. We used the pre-trained BioBERT (Bidirectional Encoder Representations from Transformers for Biomedical Text Mining) model and trained it on our biomaterial abstracts dataset. This technique is known as fine-tuning. BioBERT is a domain-specific language representation model pre-trained on large-scale biomedical corpora.
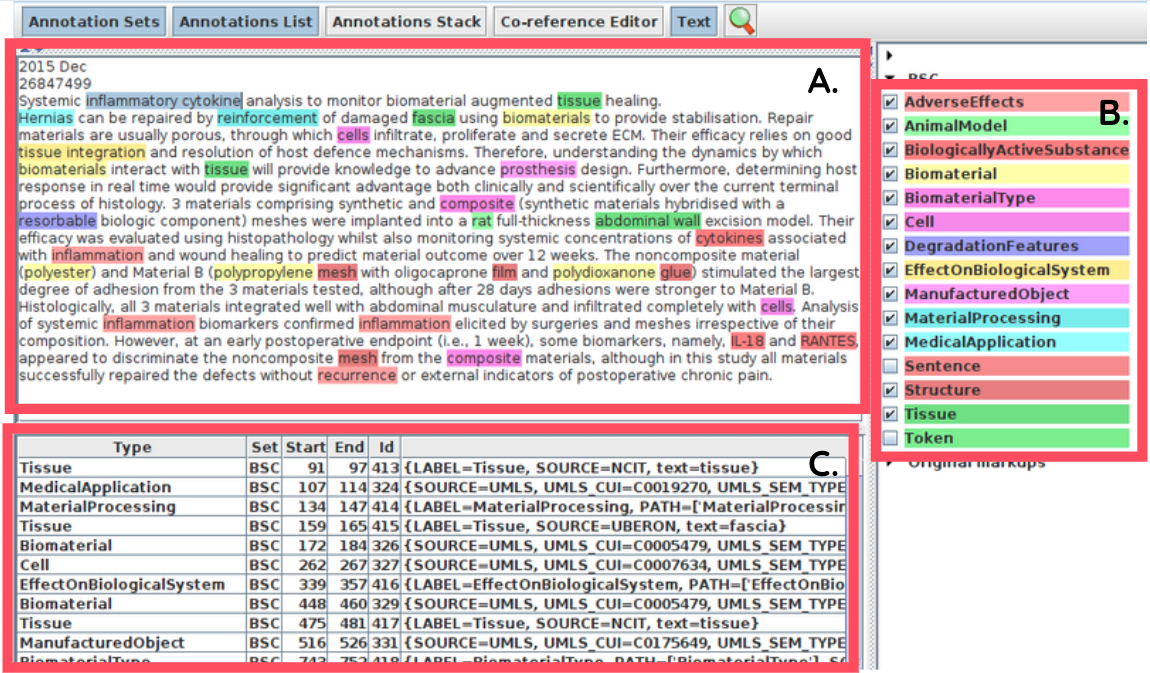
Automated text annotation is a Natural Language Processing (NLP) technique that identifies and extracts relevant concepts hidden within large collections of textual data through computational approaches. In DEBBIE, annotations are performed using the Biomaterials Annotator, an instrument combining several open lexical resource and built using the General Architecture of Text Engineering (GATE) software and the Stanford Core Natural Language Processing (CoreNLP) framework. Below is an example of an annotated abstract in GATE, with identified terms labeled with their respective categories.
| Category | Definition | Examples |
|---|---|---|
| Adverse Effects | An unfavorable or unintended disease, sign, or symptom (including an abnormal laboratory finding) that is temporally associated with the use of a medical device or biomaterial | Cytotoxicity, Inflammatory reaction, Abscesses |
| Associated Biological Process | A cellular or biological process that the manufactured object is designed to cause or support, or is measured to affect | Adipogenesis, Angiogenesis, Cell attachment |
| Biomaterial | A non-drug raw material or substance suitable for inclusion in systems which augment or replace the function of bodily tissues or organs | Polydioxanone, Polyglycolide, Hydroxyapatite |
| Biomaterial Types | Classification or nature of biomaterials | Polymer, Ceramic, Metal |
| Biological Active Substance | Substance included in a manufactured object in order to impart a biological activity | Collagen, Heparin, RGD |
| Cell | The reported cell line or primary cell type | Fibroblast, Type II Pneumocyte, Osteocyte |
| Effect on Biological System | The effect associated with manufactured object in a specific test system (cells, tissue or organism) | Biocompatibility, Cytocompatibility, Immunomodulatory |
| Manufactured Object | A physical object created by hand or machine | Experimental scaffold, Medical device, Surgical implant |
| Manufactured Object Component | A part, region or component referred to as a distinct unit, such as a surface or a layer | Core, Shell, Coat |
| Material Processing | A planned process which results in physical changes in a specified input material | Biofabrication, Coating, Knitting |
| Manufactured Object Features | Characteristics inherent or given during processing to a manufactured object or its components | Geometry, Mechanical Property, Physical Property |
| Medical Application, Disease or Condition | Intended use, context, function or outcome of the manufactured object | Artificial organs, Encapsulation, Diabetes, Injury |
| Research Technique | The reported laboratory technique used in an experimental study | Scanning electron microscope, High Performance Liquid Chromatography |
| Species | The species and /or breed used in the study | Rat, Rabbit, Mouse |
| Structure | The configuration, form or texture associated with a manufactured object or its components | Fiber, Gel, Mesh |
| Tissue | A tissue or an organ mentioned in the study as the target or test system for the biomaterial object or medical device | Lung epithelium, Nerve plexus, Elastic cartilage tissue |
Corvi, J. O., McKitrick, A., Fernández, J. M., Fuenteslópez, C. V., Gelpí, J. L., Ginebra, M. P., Capella-Gutierrez, S., & Hakimi, O. (2023). DEBBIE: The Open Access Database of Experimental Scaffolds and Biomaterials Built Using an Automated Text Mining Pipeline. Advanced healthcare materials, 12(25), e2300150. https://doi.org/10.1002/adhm.202300150
Corvi, J., Fuenteslópez, C., Fernández, J., Gelpi, J., Ginebra, M.-P., Capella-Guitierrez, S., Hakimi, O. The biomaterials annotator: a systemfor ontology-based concept annotation of biomaterials text. In:Proceedings of the Second Workshop on Scholarly DocumentProcessing, pp. 36–48. Association for Computational Linguistics,Online (2021). https://www.aclweb.org/anthology/2021.sdp-1.5
Hakimi, O., Gelpi, J., Krallinger, M., Curi, F., Repchevsky, D., Ginebra, M.-P. The devices, experimental scaffolds, and biomaterials ontology (deb): A tool for mapping, annotation, and analysis of biomaterials’ data. Adv. Funct. Mater. (2020)

This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 751277.