
The Database of Experimental Biomaterials and their Biological Effect (DEBBIE) was an EU Horizon 2020 funded project to develop an open access database of biomaterials automatically curated from the scientific literature. DEBBIE was designed to facilitate a more efficient access to the large literature in the field, generate a comprehensive map of research activity and findings, and enable evidence-based selection of materials for medical applications.
Beyond the development of database, the project was dedicated to the creation, adaptation and optimization of text mining tools for the biomaterials domain. These tools, including the DEBBIE retrieval and annotation pipeline (see illustration below), are openly available for download and use through the project’s GitHub repositories.
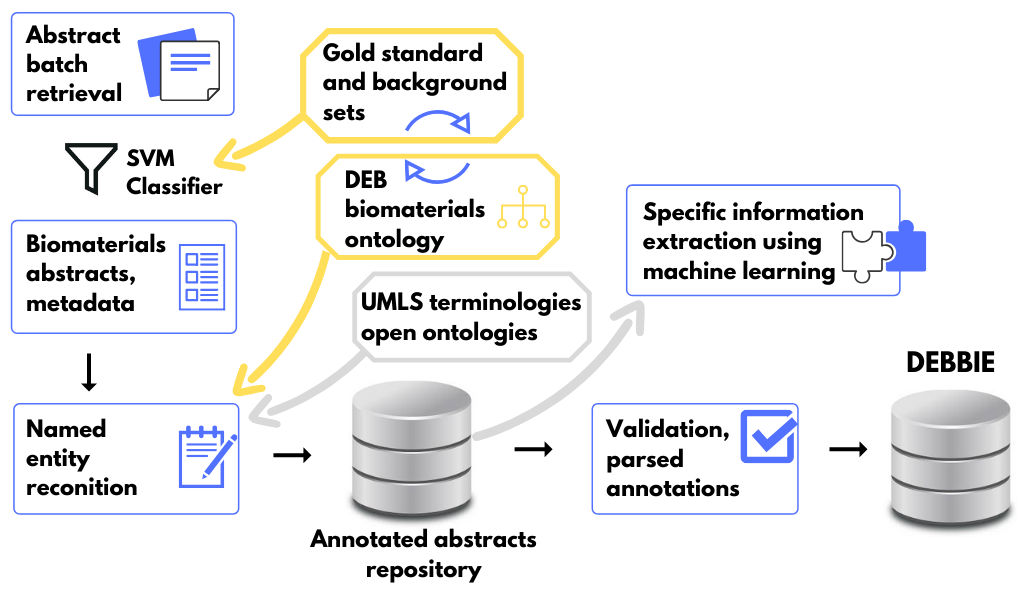
The Devices, Experimental scaffolds and Biomaterials Ontology (DEB) is an open resource for organizing information about biomaterials, their design, manufacture and biological testing. It was developed using text analysis for identifying ontology terms from a biomaterials gold standard corpus, systematically curated to represent the domain’s lexicon. Topics covered by DEB were validated by members of the biomaterials research community.
DEB may be used
for searching terms, performing annotations for machine learning applications, standardized meta-data indexing
and other cross-disciplinary data exploitation.
If you are a biomaterials scientist, we encourage and welcome your input to this effort! You can flag new terms, definitions and errors through
the 'issues' tab in DEB's GitHub repository.
In addition, below you can find the static and dynamic visualization of the ontology.
An experimental version of DEBBIE is now openly available for browsing. It is still under development, but we consider it a minimal viability product and are welcoming the feedback and comments of the research community.
The current version of the database contains abstracts automatically retrieved from PubMed, which were classified as ‘relevant’ to the areas of implants, medical devices, and experimental scaffolds. The abstracts were then annotated using our novel data model which includes multiple lexical resources. The annotations are finally stored a NoSQL database, with each of the annotated concept labelled by one or more of the following categories:
Users can access the information with a simple and highly institutive keyword search through the following link:

Could data analysis prevent unnecessary suffering? Lessons to learn from the vaginal mesh scandal.
Medium, December 2019
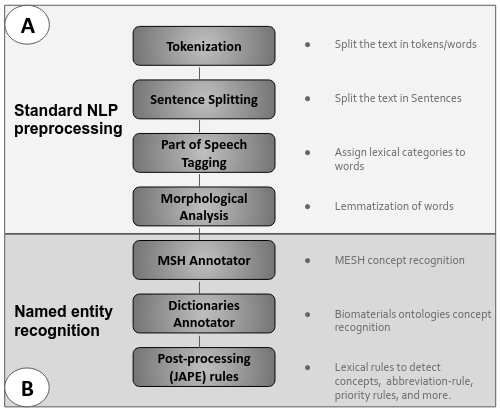
The Biomaterials Annotator: a system for ontology-based concept annotation of biomaterials text
Association for Computational Linguistics, June 2021

Project Leader
Osnat is a Bosch-Aymerich tenure tracked fellow at the Universitat Internacional de Catalunya and a visitor to the Barcelona Supercomputing Center

Bioinformatics Researcher
Javi is a Research Engineer at the Barcelona Supercomputing Center

Research Assistant
Austin is a Masters Student in Bioinformatics for Health Sciences at Universitat Pompeu Fabra (UPF) in Barcelona

Collaborator
Carla is a DPhil Student studying Engineering Science at the University of Oxford

Maria-Pau leads the Department of Materials Science and Engineering at the Universitat Politècnica de Catalunya

Martin leads the Text Mining Unit at the Barcelona Supercomputing Center

Josep leads the Computational Team for the National Institute of Bioinformatics at the Barcelona Supercomputing Center

Alexandre leads the Bioinformatics and Biomedical Signals Laboratory (B2S) at the Universitat Politècnica de Catalunya
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 751277